|
VACCINE DEVELOPMENT - A BIOINFORMATICS APPROACH |
|
|
Kumaran K. |
|
|
Guest Lecture |
|
|
With the advent of various automation strategies and robotic techniques in the field of Molecular Biology, the amount of data that is flooding is enormous. This revolution in the field of Molecular Biology in the last decade has brought in various disciplines like Physics, chemistry, Statistics and Computer Science to give birth to a new field called Bioinformatics. |
|
|
What is Bioinformatics ? |
|
|
Bio - Informatics: is conceptualizing biology in terms of molecules (in the sense of physical chemistry) and applying "Informatics techniques" (derived from disciplines such as Applied Maths, Computer Science and Statistics) to understand and organize the information associated with these molecules, on a large scale. In short Bioinformatics is a management of information system for Molecular Biology and has many practical applications. |
|
|
Infectious diseases remain a major cause of death and disabilities worldwide. Though there are several strategies developed to combat these infectious diseases, biologists have focused on vaccination as the best defense against numerous bacterial and viral pathogens. However, vaccines are still not available against many major pathogens such as Mycobacterium tuberculosis, Gonococcus, Helicobacter pylori, Shigella, etc. Vaccine development for obligate pathogens with well-defined virulence mechanisms has progressed well, but the bacteria in the focus of current efforts possess more complex pathogenesis. Thus, in spite of a wealth of antigens that have been identified and shown to be protective in animal studies, few fulfill all the requirements of a viable vaccine candidate to control human disease. |
|
|
|
|
|
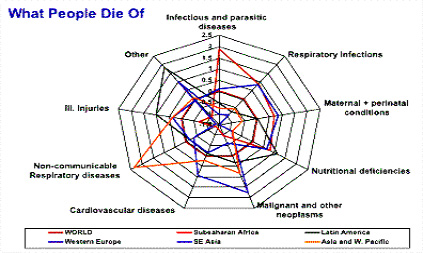
Fig : 1 WHO report on death due to various diseases |
|
|
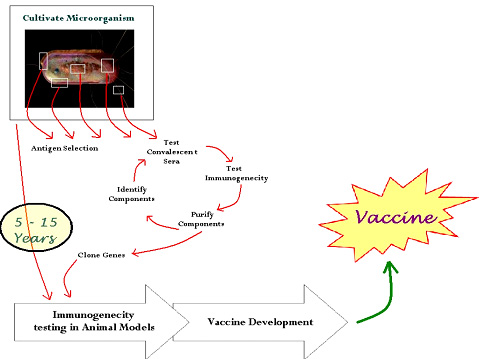
In conventional vaccinology, the identification of protective antigens is based on purification of some of the molecules produced by a pathogen and analysis of their recognition by antibodies or immune cells. Alternatively, each antigen could also be tested for its ability to induce protective immunity. Although successful in several cases, this method is time-consuming and failed to provide a solution for many human pathogens. This approach suffers many practical limitations. In particular, it relies on the identification of abundant or immunodominant antigens, that could not be the best target for induction of protection against the pathogen. It is also limited to genes that are expressed upon in vitro culture of the pathogen. The time taken comes around 10 - 15 years. The availability of complete genome sequences of pathogens has dramatically changed the scope for developing improved and novel vaccines by increasing the speed of target identification. |
|
|
|
|
|
Fig 2 : Schematic representation of Conventional Method for Vaccine Development |
|
|
To date, approximately 155 bacterial genome sequences have been determined, and more than 478 are to be completed in the near future (http://genomesonline.org). This wealth of information allows one skilled in the art of bioinformatics to search for genes of interest, a process commonly referred to as Genomic mining. Genome sequences of bacterial pathogens contain an average of 2700 genes, thus appropriate selection criteria must be applied to reduce the number of proteins for empirical testing. |
|
|
The availability of whole genome sequences of most important pathogens has dramatically changed the way we can identify potential target for vaccine development. Indeed, computer analysis can now be used to predict antigens that could be potential vaccine candidates. Although this approach is limited to proteins and thus cannot predict all other antigens such as polysaccharides or glycolipids, it has the major advantage to be not limited to the structural components of the pathogens. |
|
|
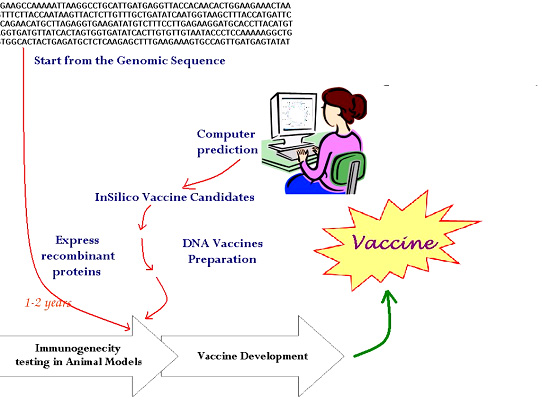
This genome-based, systematic search for vaccine candidates has been termed 'Reverse Vaccinology', and its validity was first confirmed by the identification of protective antigens from Meningococcus serotype B and later from Pneumococcus. Ninety-one novel surface-exposed proteins were identified and 28 of these proteins were shown to have bactericidal activity. Amongst these new antigens, a protein NadA was shown to be present in 52 out of 53 strains of hypervirulent meningococcus B. This antigen was recently demonstrated to be protective in the infant rat model and this could be a promising target for vaccine development. This genome-wide in silico prediction typically targets approximately 10-25% of all genome-encoded proteins and necessitates high throughput cloning and recombinant protein expression for target validation. |
|
|
|
|
|
Fig 3 : Schematic representation of the Genome - Based method for Vaccine Development (Reverse Vaccinology) |
|
|
This approach can be applied on any pathogen whether it is Bacteria or Virus. The concept of Reverse Vaccinology is novel for viruses. The approach to Vaccines against disease caused by Viruses has been conventional, only structural antigens have been taken into account. Recent studies indicate that the genome could provide information about potential antigens, which are either not part of the final viral particle or are present in very low quantities that they could not be purified and used as antigens by conventional strategies. Thus, applying the concept of Reverse Vaccinology can provide insights in finding potential antigens for Viruses. |
|
|
Several recent studies have shown that genomics-based technologies can add considerable value to the vaccine field. However, despite the fast and efficient target identification, vaccinologists are still faced with the slow and laborious validation steps that are performed using conventional techniques. Improvements and the combined use of the antigen discovery technologies, each associated with distinct limitations, could reduce this validation burden and increase the success rate in vaccine development, by selecting the most relevant candidates that fulfill most, if not all of the criteria of useful vaccine antigens. The expectation for these technologies is to identify vaccine targets that can protect humans from infectious diseases. In addition, these high-throughput genomics-based approaches will contribute to the better understanding of microbial pathogenesis, host-pathogen interactions and protective immune responses. This knowledge will help vaccinologists to prioritize antigens and establish the rational design of a new generation of recombinant vaccines. |
|
|
The genomics-based technologies not only helps in accelerating vaccine discovery, but also reduces the time and cost which could be a major beneficial effect for the developing countries to combat various infectious diseases. |
|